
Illustration of approximate non-negative matrix factorization: the matrix
V is represented by the two smaller matrices
W and
H, which, when multiplied, approximately reconstruct
V.
- NMF redirects here. For the bridge convention, see new minor forcing.
Non-negative matrix factorization (NMF), also non-negative matrix approximation[1][2] is a group of algorithms in multivariate analysis and linear algebra where a matrix V is factorized into (usually) two matrices W and H, with the property that all three matrices have no negative elements. This non-negativity makes the resulting matrices easier to inspect. Since the problem is not exactly solvable in general, it is commonly approximated numerically.
NMF finds applications in such fields as computer vision, document clustering,[1] chemometrics and recommender systems.[3]
Contents
[hide] - 1 History
- 2 Background
- 3 Types
- 3.1 Approximate non-negative matrix factorization
- 3.2 Different cost functions and regularizations
- 4 Algorithms
- 5 Relation to other techniques
- 6 Uniqueness
- 7 Applications
- 7.1 Text mining
- 7.2 Spectral data analysis
- 7.3 Scalable Internet distance prediction
- 7.4 Non-stationary speech denoising
- 7.5 Bioinformatics
- 8 Current research
- 9 See also
- 10 Sources and external links
- 10.1 Notes
- 10.2 Others
- 10.3 Software
History[edit]
In chemometrics non-negative matrix factorization has a long history under the name "self modeling curve resolution".[4] In this framework the vectors in the right matrix are continuous curves rather than discrete vectors. Also early work on non-negative matrix factorizations was performed by a Finnish group of researchers in the middle of the 1990s under the name positive matrix factorization.[5][6] It became more widely known as non-negative matrix factorization after Lee and Seung investigated the properties of the algorithm and published some simple and useful algorithms for two types of factorizations.[7][8]
Background[edit]
Let matrix 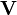 be the product of the matrices
be the product of the matrices  and
and  such that:
such that:
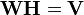
Matrix multiplication can be implemented as linear combinations of column vectors in  with coefficients supplied by cell values in
with coefficients supplied by cell values in  . Each column in
. Each column in 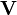 can be computed as follows:
can be computed as follows:
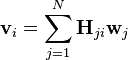
where: 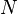 is the number of columns in
is the number of columns in 
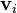 is the
is the  column vector of the product matrix
column vector of the product matrix 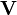
 is the cell value in the
is the cell value in the 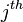 row and
row and  column of the matrix
column of the matrix 
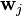 is the
is the 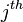 column of the matrix
column of the matrix 
When multiplying matrices, the dimensions of the factor matrices may be significantly lower than those of the product matrix and it's this property that forms the basis of NMF. NMF generates factors with significantly reduced dimensions compared to the original matrix. For example, if 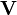 is an
is an 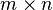 matrix, then
matrix, then  is an
is an  matrix, and
matrix, and  is a
is a  matrix. Here
matrix. Here 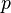 can be significantly less than both
can be significantly less than both 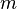 and
and  .
.
Here's an example based on a text-mining application:
- Let the input matrix (the matrix to be factored) be
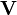 with 10000 rows and 500 columns where words are in rows and documents are in columns. In other words, we have 500 documents indexed by 10000 words. It follows that a column vector
with 10000 rows and 500 columns where words are in rows and documents are in columns. In other words, we have 500 documents indexed by 10000 words. It follows that a column vector  in
in 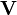 represents a document.
represents a document. - Assume we ask the algorithm to find 10 features in order to generate a features matrix
 with 10000 rows and 10 columns and a coefficients matrix
with 10000 rows and 10 columns and a coefficients matrix  with 10 rows and 500 columns.
with 10 rows and 500 columns. - The product of
 and
and  is a matrix with 10000 rows and 500 columns, the same shape as the input matrix
is a matrix with 10000 rows and 500 columns, the same shape as the input matrix 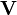 and, if the factorization worked, also a reasonable approximation to the input matrix
and, if the factorization worked, also a reasonable approximation to the input matrix 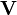 .
. - From the treatment of matrix multiplication above it follows that each column in the product matrix
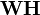 is a linear combination of the 10 column vectors in the features matrix
is a linear combination of the 10 column vectors in the features matrix  with coefficients supplied by the coefficients matrix
with coefficients supplied by the coefficients matrix  .
.
This last point is the basis of NMF because we can consider each original document in our example as being built from a small set of hidden features. NMF generates these features.
It's useful to think of each feature (column vector) in the features matrix  as a document archetype comprising a set of words where each word's cell value defines the word's rank in the feature: The higher a word's cell value the higher the word's rank in the feature. A column in the coefficients matrix
as a document archetype comprising a set of words where each word's cell value defines the word's rank in the feature: The higher a word's cell value the higher the word's rank in the feature. A column in the coefficients matrix  represents an original document with a cell value defining the document's rank for a feature. This follows because each row in
represents an original document with a cell value defining the document's rank for a feature. This follows because each row in  represents a feature. We can now reconstruct a document (column vector) from our input matrix by a linear combination of our features (column vectors in
represents a feature. We can now reconstruct a document (column vector) from our input matrix by a linear combination of our features (column vectors in  ) where each feature is weighted by the feature's cell value from the document's column in
) where each feature is weighted by the feature's cell value from the document's column in  .
.
Approximate non-negative matrix factorization[edit]
Usually the number of columns of W and the number of rows of H in NMF are selected so the product WH will become an approximation to V. The full decomposition of V then amounts to the two non-negative matrices W and H as well as a residual U, such that: V = WH + U. The elements of the residual matrix can either be negative or positive.
When W and H are smaller than V they become easier to store and manipulate. Another reason for factorizing V into smaller matrices W and H, is that if one is able to approximately represent the elements of V by significantly less data, then one has to infer some latent structure in the data.
Different cost functions and regularizations[edit]
There are different types of non-negative matrix factorizations. The different types arise from using different cost functions for measuring the divergence between V and WH and possibly by regularization of the W and/or H matrices.[1]
Two simple divergence functions studied by Lee and Seung are the squared error (or Frobenius norm) and an extension of the Kullback-Leibler divergence to positive matrices (the original Kullback-Leibler divergence is defined on probability distributions). Each divergence leads to a different NMF algorithm, usually minimizing the divergence using iterative update rules.
The factorization problem in the squared error version of NMF may be stated as: Given a matrix 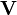 find nonnegative matrices W and H that minimize the function
find nonnegative matrices W and H that minimize the function
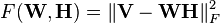
Another type of NMF for images is based on the total variation norm.[9]
When L1 regularization is added to NMF with the mean squared error cost function, the resulting problem is also called non-negative sparse coding due to the similarity to the sparse coding problem.[10]
Algorithms[edit]
There are several ways in which the W and H may be found: Lee and Seung's multiplicative update rule [8] has been a popular method due to the simplicity of implementation. Since then, a few other algorithmic approaches have been developed.
Some successful algorithms are based on alternating non-negative least squares, in which the nonnegativity constrained least squares subproblems are iteratively solved. Specific approaches include the projected gradient descent methods,[11][12] the active-set method,[13][3] and the block principal pivoting method[14] among several others.
The currently available algorithms are sub-optimal as they can only guarantee finding a local minimum, rather than a global minimum of the cost function. A provably optimal algorithm is unlikely in the near future as the problem has been shown to generalize the k-means clustering problem which is known to be computationally difficult (NP-complete).[15] However, as in many other data mining applications a local minimum may still prove to be useful.
Arora, Ge, Kannan, and Moitra (2012) have given an algorithm for exact NMF that provably runs in polynomial-time (i.e., always halts with the correct answer) if one of the factors W satisfies the separability condition. This condition is observed to hold in factorizations empirically found in many settings. They also give a more precise analysis of the complexity of NMF as a function of the dimension of W and H. [16]
Relation to other techniques[edit]
In Learning the parts of objects by non-negative matrix factorization Lee and Seung proposed NMF mainly for parts-based decomposition of images. It compares NMF to vector quantization and principal component analysis, and shows that although the three techniques may be written as factorizations, they implement different constraints and therefore produce different results.
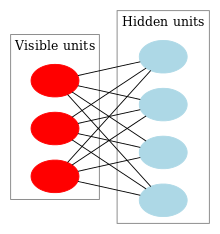
NMF as a probabilistic graphical model: visible units (
V) are connected to hidden units (
H) through weights
W, so that
V is
generated from a probability distribution with mean

.
[7]:5It was later shown that some types of NMF are an instance of a more general probabilistic model called "multinomial PCA".[17] When NMF is obtained by minimizing the Kullback–Leibler divergence, it is in fact equivalent to another instance of multinomial PCA, probabilistic latent semantic analysis,[18] trained by maximum likelihood estimation. That method is commonly used for analyzing and clustering textual data and is also related to the latent class model.
It has been shown [19][20] NMF is equivalent to a relaxed form of K-means clustering: matrix factor W contains cluster centroids and H contains cluster membership indicators, when using the least square as NMF objective. This provides theoretical foundation for using NMF for data clustering.
NMF extends beyond matrices to tensors of arbitrary order.[21][22][23] This extension may be viewed as a non-negative version of, e.g., the PARAFAC model.
Other extensions of NMF include joint factorisation of several data matrices and tensors where some factors are shared. Such models are useful for sensor fusion and relational learning.[24]
NMF is an instance of the nonnegative quadratic programming (NQP) as well as many other important problems including the support vector machine (SVM). However, SVM and NMF are related at a more intimate level than that of NQP, which allows direct application of the solution algorithms developed for either of the two methods to problems in both domains.[25]
Uniqueness[edit]
The factorization is not unique: A matrix and its inverse can be used to transform the two factorization matrices by, e.g.,[26]
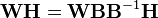
If the two new matrices 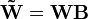 and
and  are non-negative they form another parametrization of the factorization.
are non-negative they form another parametrization of the factorization.
The non-negativity of  and
and 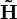 applies at least if B is a non-negative monomial matrix. In this simple case it will just correspond to a scaling and a permutation.
applies at least if B is a non-negative monomial matrix. In this simple case it will just correspond to a scaling and a permutation.
More control over the non-uniqueness of NMF is obtained with sparsity constraints.[27]
Applications[edit]
Text mining[edit]
NMF can be used for text mining applications. In this process, a document-term matrix is constructed with the weights of various terms (typically weighted word frequency information) from a set of documents. This matrix is factored into a term-feature and afeature-document matrix. The features are derived from the contents of the documents, and the feature-document matrix describes data clusters of related documents.
One specific application used hierarchical NMF on a small subset of scientific abstracts from PubMed.[28] Another research group clustered parts of the Enron email dataset[29] with 65,033 messages and 91,133 terms into 50 clusters.[30] NMF has also been applied to citations data, with one example clustering Wikipedia articles and scientific journals based on the outbound scientific citations in Wikipedia.[31]
Arora, Ge and Moitra (2012) have given polynomial-time algorithms to learn topic models using NMF. The algorithm assumes that the topic matrix satisfies a separability condition that is often found to hold in these settings. [32]
Spectral data analysis[edit]
NMF is also used to analyze spectral data; one such use is in the classification of space objects and debris.[33]
Scalable Internet distance prediction[edit]
NMF is applied in scalable Internet distance (round-trip time) prediction. For a network with 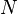 hosts, with the help of NMF, the distances of all the
hosts, with the help of NMF, the distances of all the 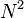 end-to-end links can be predicted after conducting only
end-to-end links can be predicted after conducting only  measurements. This kind of method was firstly introduced in Internet Distance Estimation Service (IDES).[34] Afterwards, as a fully decentralized approach, Phoenix network coordinate system [35] is proposed. It achieves better overall prediction accuracy by introducing the concept of weight.
measurements. This kind of method was firstly introduced in Internet Distance Estimation Service (IDES).[34] Afterwards, as a fully decentralized approach, Phoenix network coordinate system [35] is proposed. It achieves better overall prediction accuracy by introducing the concept of weight.
Non-stationary speech denoising[edit]
Speech denoising has been a long lasting problem in audio signal processing. There are lots of algorithms for denoising if the noise is stationary. For example, the Wiener filter is suitable for additive Gaussian noise. However, if the noise is non-stationary, the classical denoising algorithms usually have poor performance because the statistical information of the non-stationary noise is difficult to estimate. Schmidt et al.[36] use NMF to do speech denoising under non-stationary noise, which is completely different than classical statistical approaches.The key idea is that clean speech signal can be sparsely represented by a speech dictionary, but non-stationary noise cannot. Similarly, non-stationary noise can also be sparsely represented by a noise dictionary, but speech cannot.
The algorithm for NMF denoising goes as follows. Two dictionaries, one for speech and one for noise, need to be trained offline. Once a noisy speech is given, we first calculate the magnitude of the Short-Time-Fourier-Transform. Second, separate it into two parts via NMF, one can be sparsely represented by the speech dictionary, and the other part can be sparsely represented by the noise dictionary. Third, the part that is represented by the speech dictionary will be the estimated clean speech.
Bioinformatics[edit]
NMF has been successfully applied to bioinformatics.[37][38]
Current research[edit]
Current[when?] research in nonnegative matrix factorization includes, but not limited to,
(1) Algorithmic: searching for global minima of the factors and factor initialization.[39]
(2) Scalability: how to factorize million-by-billion matrices, which are commonplace in Web-scale data mining, e.g., see Distributed Nonnegative Matrix Factorization (DNMF)[40]
(3) Online: how to update the factorization when new data comes in without recomputing from scratch.
See also[edit]
Sources and external links[edit]
- ^ Jump up to:a b c Inderjit S. Dhillon, Suvrit Sra (2005). "Generalized Nonnegative Matrix Approximations with Bregman Divergences"(PDF). NIPS.
- Jump up^ Tandon, Rashish; Suvrit Sra (September 2010). Sparse nonnegative matrix approximation: new formulations and algorithms. TR.
- ^ Jump up to:a b Rainer Gemulla, Erik Nijkamp, Peter J Haas, Yannis Sismanis (2011). "Large-scale matrix factorization with distributed stochastic gradient descent". Proc. ACM SIGKDD Int'l Conf. on Knowledge discovery and data mining. pp. 69–77.
- Jump up^ William H. Lawton; Edward A. Sylvestre (August 1971). "Self modeling curve resolution". Technometrics 13 (3): 617+.
- Jump up^ P. Paatero, U. Tapper (1994). "Positive matrix factorization: A non-negative factor model with optimal utilization of error estimates of data values". Environmetrics 5 (2): 111–126. doi:10.1002/env.3170050203.
- Jump up^ Pia Anttila, Pentti Paatero, Unto Tapper, Olli Järvinen (1995). "Source identification of bulk wet deposition in Finland by positive matrix factorization". Atmospheric Environment 29 (14): 1705–1718. doi:10.1016/1352-2310(94)00367-T.
- ^ Jump up to:a b Daniel D. Lee and H. Sebastian Seung (1999). "Learning the parts of objects by non-negative matrix factorization". Nature401 (6755): 788–791. doi:10.1038/44565. PMID 10548103.
- ^ Jump up to:a b Daniel D. Lee and H. Sebastian Seung (2001). "Algorithms for Non-negative Matrix Factorization". Advances in Neural Information Processing Systems 13: Proceedings of the 2000 Conference. MIT Press. pp. 556–562.
- Jump up^ Taiping Zhanga, Bin Fang, Weining Liu, Yuan Yan Tang, Guanghui He and Jing Wen (June 2008). "Total variation norm-based nonnegative matrix factorization for identifying discriminant representation of image patterns". Neurocomputing 71 (10–12): 1824–1831. doi:10.1016/j.neucom.2008.01.022.
- Jump up^ Hoyer, Patrik O. (2002). "Non-negative sparse coding". Proc. IEEE Workshop on Neural Networks for Signal Processing.
- Jump up^ Chih-Jen Lin (October 2007). "Projected Gradient Methods for Non-negative Matrix Factorization" (PDF). Neural Computation19 (10).
- Jump up^ Chih-Jen Lin (November 2007). "On the Convergence of Multiplicative Update Algorithms for Nonnegative Matrix Factorization".IEEE Transactions on Neural Networks 18 (6): 1589–1596. doi:10.1109/TNN.2007.895831.
- Jump up^ Hyunsoo Kim and Haesun Park (2008). "Nonnegative Matrix Factorization Based on Alternating Nonnegativity Constrained Least Squares and Active Set Method". SIAM Journal on Matrix Analysis and Applications 30 (2): 713–730.
- Jump up^ Jingu Kim and Haesun Park (2011). "Fast Nonnegative Matrix Factorization: An Active-set-like Method and Comparisons".SIAM Journal on Scientific Computing 33 (6): 3261–3281.
- Jump up^ Ding, C. and He, X. and Simon, H.D., (2005). "On the equivalence of nonnegative matrix factorization and spectral clustering".Proc. SIAM Data Mining Conf 4: 606–610.
- Jump up^ Sanjeev Arora and Rong Ge and Ravi Kannan and Ankur Moitra ({2012}). "Computing a Nonnegative Matrix Factorization ---Provably.". ACM STOC.
- Jump up^ Wray Buntine (2002). "Variational Extensions to EM and Multinomial PCA" (PDF). Proc. European Conference on Machine Learning (ECML-02). LNAI 2430. pp. 23–34.
- Jump up^ Eric Gaussier and Cyril Goutte (2005). "Relation between PLSA and NMF and Implications" (PDF). Proc. 28th international ACM SIGIR conference on Research and development in information retrieval (SIGIR-05). pp. 601–602.
- Jump up^ Chris Ding, Xiaofeng He, and Horst D. Simon (2005). "On the Equivalence of Nonnegative Matrix Factorization and Spectral Clustering". Proc. SIAM Int'l Conf. Data Mining, pp. 606-610. May 2005
- Jump up^ Ron Zass and Amnon Shashua (2005). "A Unifying Approach to Hard and Probabilistic Clustering". International Conference on Computer Vision (ICCV) Beijing, China, Oct., 2005.
- Jump up^ Pentti Paatero (1999). "The Multilinear Engine: A Table-Driven, Least Squares Program for Solving Multilinear Problems, including the n-Way Parallel Factor Analysis Model". Journal of Computational and Graphical Statistics 8 (4): 854–888.doi:10.2307/1390831. JSTOR 1390831.
- Jump up^ Max Welling and Markus Weber (2001). "Positive Tensor Factorization". Pattern Recognition Letters 22 (12): 1255–1261.doi:10.1016/S0167-8655(01)00070-8.
- Jump up^ Jingu Kim and Haesun Park (2012). "Fast Nonnegative Tensor Factorization with an Active-set-like Method". High-Performance Scientific Computing: Algorithms and Applications. Springer. pp. 311–326.
- Jump up^ Kenan Yilmaz and A. Taylan Cemgil and Umut Simsekli ({2011}). "Generalized Coupled Tensor Factorization". NIPS.
- Jump up^ Vamsi K. Potluru and Sergey M. Plis and Morten Morup and Vince D. Calhoun and Terran Lane (2009). "Efficient Multiplicative updates for Support Vector Machines". Proceedings of the 2009 SIAM Conference on Data Mining (SDM). pp. 1218–1229.
- Jump up^ Wei Xu, Xin Liu & Yihong Gong (2003). "Document clustering based on non-negative matrix factorization". Proceedings of the 26th annual international ACM SIGIR conference on Research and development in information retrieval. New York: Association for Computing Machinery. pp. 267–273.
- Jump up^ Julian Eggert, Edgar Körner, "Sparse coding and NMF", Proceedings. 2004 IEEE International Joint Conference on Neural Networks, 2004., pp. 2529-2533, 2004.
- Jump up^ Nielsen, Finn Årup; Balslev, Daniela; Hansen, Lars Kai (September 2005). "Mining the posterior cingulate: segregation between memory and pain components". NeuroImage 27 (3): 520–522. doi:10.1016/j.neuroimage.2005.04.034.PMID 15946864.
- Jump up^ Cohen, William (2005-04-04). "Enron Email Dataset". Retrieved 2008-08-26.
- Jump up^ Berry, Murray; Browne, Murray (October 2005). "Email Surveillance Using Non-negative Matrix Factorization". Computational and Mathematical Organization Theory 11 (3): 249–264. doi:10.1007/s10588-005-5380-5.
- Jump up^ Nielsen, Finn Årup (2008). "Clustering of scientific citations in Wikipedia". Wikimania.
- Jump up^ Sanjeev Arora and Rong Ge and Ankur Moitra ({2012}). "Learning Topic Models---Going beyond SVD.". arxiv.
- Jump up^ Michael W. Berry, et al. (June 2006). Algorithms and Applications for Approximate Nonnegative Matrix Factorization.
- Jump up^ Yun Mao, Lawrence Saul and Jonathan M. Smith (December 2006). "IDES: An Internet Distance Estimation Service for Large Networks". IEEE Journal on Selected Areas in Communications 24 (12): 2273–2284. doi:10.1109/JSAC.2006.884026.
- Jump up^ Yang Chen, Xiao Wang, Cong Shi, and et al. (December 2011). "Phoenix: A Weight-based Network Coordinate System Using Matrix Factorization" (PDF). IEEE Transactions on Network and Service Management 8 (4): 334–347.
- Jump up^ Schmidt, M.N., J. Larsen, and F.T. Hsiao. (2007). "Wind noise reduction using non-negative sparse coding", Machine Learning for Signal Processing, IEEE Workshop on, 431–436
- Jump up^ Devarajan, K. (2008). "Nonnegative Matrix Factorization: An Analytical and Interpretive Tool in Computational Biology". PLoS Computational Biology 4 (7).
- Jump up^ Hyunsoo Kim and Haesun Park (2007). "Sparse non-negative matrix factorizations via alternating non-negativity-constrained least squares for microarray data analysis". Bioinformatics 23 (12): 1495–1502. doi:10.1093/bioinformatics/btm134.PMID 17483501.
- Jump up^ C. Boutsidis and E. Gallopoulos (April 2008). "SVD based initialization: A head start for nonnegative matrix factorization". Pattern Recognition 41 (4): 1350–1362. doi:10.1016/j.patcog.2007.09.010.
- Jump up^ Chao Liu, Hung-chih Yang, Jinliang Fan, Li-Wei He, and Yi-Min Wang (April 2010). "Distributed Nonnegative Matrix Factorization for Web-Scale Dyadic Data Analysis on MapReduce". Proceedings of the 19th International World Wide Web Conference.
Software[edit]