Manifold alignment is a class of machine learning algorithms that produce projections between sets of data, given that the original data sets lie on a common manifold. The concept was first introduced as such by Ham, Lee, and Saul in 2003,[1] adding a manifold constraint to the general problem of correlating sets of high-dimensional vectors.[2]
Contents
[hide] - 1 Overview
- 2 Inter-data correspondences
- 3 One-step vs Two-step alignment
- 4 Instance-level vs Feature-level projections
- 5 Applications
- 6 References
- 7 Further reading
Overview[edit]
Manifold alignment assumes that disparate data sets produced by similar generating processes will share a similar underlying manifold representation. By learning projections from each original space to the shared manifold, correspondences are recovered and knowledge from one domain can be transferred to another. Most manifold alignment techniques consider only two data sets, but the concept extends to arbitrarily many initial data sets.
Consider the case of aligning two data sets, 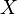 and
and 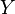 , with
, with 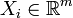 and
and 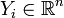 .
.
Manifold alignment algorithms attempt to project both 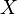 and
and 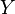 into a new d-dimensional space such that the projections both minimize distance between corresponding points and preserve the local manifold structure of the original data. The projection functions are denoted:
into a new d-dimensional space such that the projections both minimize distance between corresponding points and preserve the local manifold structure of the original data. The projection functions are denoted:

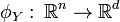
Let  represent the binary correspondence matrix between points in
represent the binary correspondence matrix between points in 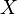 and
and 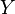 :
:

Let 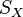 and
and 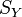 represent pointwise similarities within data sets. This is usually encoded as the heat kernel of the adjacency matrix of a k-nearest neighbor graph.
represent pointwise similarities within data sets. This is usually encoded as the heat kernel of the adjacency matrix of a k-nearest neighbor graph.
Finally, introduce a coefficient 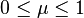 , which can be tuned to adjust the weight of the 'preserve manifold structure' goal, versus the 'minimize corresponding point distances' goal.
, which can be tuned to adjust the weight of the 'preserve manifold structure' goal, versus the 'minimize corresponding point distances' goal.
With these definitions in place, the loss function for manifold alignment can be written:
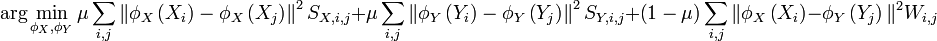
Solving this optimization problem is equivalent to solving a generalized eigenvalue problem using the graph laplacian[3] of the joint matrix, G:
![G=\left[\begin{array}{cc}\mu S_X & \left(1-\mu\right)W\\\left(1-\mu\right)W^T & \mu S_Y\end{array}\right]](http://upload.wikimedia.org/math/8/9/8/8989534ffad2047495bf081821ba98d2.png)
Inter-data correspondences[edit]
The algorithm described above requires full pairwise correspondence information between input data sets; a supervised learning paradigm. However, this information is usually difficult or impossible to obtain in real world applications. Recent work has extended the core manifold alignment algorithm to semi-supervised [4] , unsupervised [5] , and multiple-instance [6] settings.
One-step vs Two-step alignment[edit]
The algorithm described above performs a "one-step" alignment, finding embeddings for both data sets at the same time. A similar effect can also be achieved with "two-step" alignments [7] [8] , following a slightly modified procedure:
- Project each input data set to a lower-dimensional space independently, using any of a variety of dimension reduction algorithms.
- Perform linear manifold alignment on the embedded data, holding the first data set fixed, mapping each additional data set onto the first's manifold. This approach has the benefit of decomposing the required computation, which lowers memory overhead and allows parallel implementations.
Instance-level vs Feature-level projections[edit]
Manifold alignment can be used to find linear (feature-level) projections, or nonlinear (instance-level) embeddings. While the instance-level version generally produces more accurate alignments, it sacrifices a great degree of flexibility as the learned embedding is often difficult to parameterize. Feature-level projections allow any new instances to be easily embedded in the manifold space, and projections may be combined to form direct mappings between the original data representations. These properties are especially important for knowledge-transfer applications.
Applications[edit]
Manifold alignment is suited to problems with several corpora that lie on a shared manifold, even when each corpus is of a different dimensionality. Many real-world problems fit this description, but traditional techniques are not able to take advantage of all corpora at the same time. Manifold alignment also facilitates transfer learning, in which knowledge of one domain is used to jump-start learning in correlated domains.
Applications of manifold alignment include:
- Cross-language information retrieval / automatic translation[8]
- By representing documents as vector of word counts, manifold alignment can recover the mapping between documents of different languages.
- Cross-language document correspondence is relatively easy to obtain, especially from multi-lingual organizations like the European Union.
- Transfer learning of policy and state representations for reinforcement learning[8]
- Alignment of protein NMR structures[8]
References[edit]
- Jump up^ Ham, Ji Hun; Daniel D. Lee, Lawrence K. Saul (2003). "Learning high dimensional correspondences from low dimensional manifolds". Proceedings of the Twentieth International Conference on Machine Learning (ICML-2003) (Washington DC, USA).
- Jump up^ Hotelling, H (1936). "Relations between two sets of variates". Biometrika vol. 28, no. 3/4.
- Jump up^ Belkin, M; P Niyogi (2003). "Laplacian eigenmaps for dimensionality reduction and data representation". Neural Computation.
- Jump up^ Ham, Ji Hun; Daniel D. Lee, Lawrence K. Saul (2005). "Semisupervised alignment of manifolds". Proceedings of the Annual Conference on Uncertainty in Artificial Intelligence.
- Jump up^ Wang, Chang; Sridhar Mahadevan (2009). "Manifold Alignment without Correspondence". The 21st International Joint Conference on Artificial Intelligence.
- Jump up^ Wang, Chang; Sridhar Mahadevan (2011). "Heterogeneous Domain Adaptation using Manifold Alignment". The 22nd International Joint Conference on Artificial Intelligence.
- Jump up^ Lafon, Stephane; Yosi Keller, Ronald R. Coifman (2006). "Data fusion and multicue data matching by diffusion maps". IEEE transactions on Pattern Analysis and Machine Intelligence.
- ^ Jump up to:a b c d Wang, Chang; Sridhar Mahadevan (2008). "Manifold Alignment using Procrustes Analysis". The 25th International Conference on Machine Learning.
Further reading[edit]